for (pkg in c("tidyverse")) {
if (!require(pkg, character.only = TRUE)) install.packages(pkg)
}
library(tidyverse)To install and load all packages used in this chapter, run the following code:
Introduction
In practice, data rarely comes in a single, perfectly prepared table. Instead, we often have multiple data sources that need to be combined: measurements from different laboratories, master data and transaction data, or simply data spread across multiple Excel sheets. This chapter shows how to combine tables in R using various approaches.
We distinguish three fundamental approaches:
-
Stacking: Simply placing tables below each other (
bind_rows()) or next to each other (bind_cols()) - Joining: Intelligently linking tables based on common key columns
- Reshaping: Transforming data between “wide” and “long” formats
Stacking Tables
The simplest way to combine tables is “stacking” - placing tables either below or next to each other. For this purpose, we have bind_rows() and bind_cols().
Example Data
For this section, we create three small tibbles with fruit data:
fruit_1# A tibble: 2 × 2
variety price
<chr> <dbl>
1 Apple 1.2
2 Pear 1.5fruit_2# A tibble: 2 × 2
variety price
<chr> <dbl>
1 Orange 0.8
2 Banana 1.1fruit_3# A tibble: 2 × 3
variety price origin
<chr> <dbl> <chr>
1 Cherry 3.5 Germany
2 Plum 2.2 Spain Note that fruit_1 and fruit_2 have the same columns (variety and price), while fruit_3 has an additional column origin.
bind_rows()
The function bind_rows() stacks tables vertically - it adds rows. This is useful when you have data from different time periods or different sources that share the same structure.
bind_rows(fruit_1, fruit_2)# A tibble: 4 × 2
variety price
<chr> <dbl>
1 Apple 1.2
2 Pear 1.5
3 Orange 0.8
4 Banana 1.1This works as expected: the rows are simply stacked on top of each other.
Different Columns
The big advantage of bind_rows() over the base R function rbind() becomes apparent when the tables have different columns. While rbind() throws an error in this case, bind_rows() combines the tables anyway and fills missing values with NA:
bind_rows(fruit_1, fruit_3)# A tibble: 4 × 3
variety price origin
<chr> <dbl> <chr>
1 Apple 1.2 <NA>
2 Pear 1.5 <NA>
3 Cherry 3.5 Germany
4 Plum 2.2 Spain As we can see, fruit_1 had no origin column, so these values are filled with NA. This is very convenient when combining data from different sources that don’t have exactly the same columns.
Tracking Origin with .id
When combining multiple tables, we often want to know which original table each row came from. For this, there’s the .id argument:
bind_rows(
"Store_A" = fruit_1,
"Store_B" = fruit_2,
.id = "source"
)# A tibble: 4 × 3
source variety price
<chr> <chr> <dbl>
1 Store_A Apple 1.2
2 Store_A Pear 1.5
3 Store_B Orange 0.8
4 Store_B Banana 1.1Here we gave names to the tables (“Store_A”, “Store_B”) and created a new column with .id = "source" that contains these names.
Combining All Three Tables
We can also stack more than two tables at once:
bind_rows(fruit_1, fruit_2, fruit_3)# A tibble: 6 × 3
variety price origin
<chr> <dbl> <chr>
1 Apple 1.2 <NA>
2 Pear 1.5 <NA>
3 Orange 0.8 <NA>
4 Banana 1.1 <NA>
5 Cherry 3.5 Germany
6 Plum 2.2 Spain The origin column only exists for the last two rows (from fruit_3), all others get NA.
bind_cols()
The function bind_cols() combines tables horizontally - it glues columns together.
With bind_cols() there is no intelligent linking via key columns! The tables are simply “blindly” glued together side by side. This means: the rows must be in exactly the same order, and the tables must have the same number of rows.
An example:
# A tibble: 3 × 4
first_name last_name age profession
<chr> <chr> <dbl> <chr>
1 Anna Mueller 28 Physician
2 Ben Schmidt 34 Engineer
3 Clara Weber 22 Student This works because both tibbles have three rows and we know that row 1 in both tibbles belongs to the same person.
When is bind_cols() Dangerous?
bind_cols() can lead to incorrect results if the row order doesn’t match:
# A tibble: 3 × 4
first_name last_name age profession
<chr> <chr> <dbl> <chr>
1 Anna Mueller 28 Physician
2 Ben Schmidt 34 Engineer
3 Clara Weber 22 Student Here the names were sorted alphabetically, but the age data was not - Anna now gets age 28 assigned, which happened to be correct before sorting (and is coincidentally still correct), but Ben and Clara are swapped! This is a common mistake!
When Should You Use bind_cols()?
bind_cols() is safe when:
- The data comes from the same source and is guaranteed to have the same order
- You just performed multiple calculations on the same data yourself
- You verify correctness after combining
In most other cases, a join is the better choice because it links via a key column.
Joining Tables
Joins are the most powerful method for combining tables. They link tables intelligently via one or more common columns (the “keys”). This means it doesn’t matter what order the rows are in - R automatically finds the matching rows.
Example Data
For the joins, we use a different dataset: city data. We create three tibbles with different information about cities:
# Tibble 1: Six major cities in Central Europe with population
cities_europe <- tibble(
city = c("Berlin", "Hamburg", "Munich", "Copenhagen", "Amsterdam", "London"),
population_mio = c(3.9, 1.9, 1.5, 0.7, 0.9, 9.0)
)
# Tibble 2: Ten German cities with rental prices (Euro per square meter)
cities_rent <- tibble(
city = c("Berlin", "Hamburg", "Munich", "Frankfurt", "Cologne",
"Duesseldorf", "Stuttgart", "Leipzig", "Dresden", "Nuremberg"),
rent_sqm = c(18.29, 17.18, 22.64, 19.62, 15.21,
16.04, 17.26, 11.38, 7.33, 9.65)
)
# Tibble 3: The same ten German cities with additional statistics
cities_stats <- tibble(
city = c("Berlin", "Hamburg", "Munich", "Frankfurt", "Cologne",
"Duesseldorf", "Stuttgart", "Leipzig", "Dresden", "Nuremberg"),
area_km2 = c(892, 755, 310, 248, 405, 217, 207, 297, 328, 186),
green_space_pct = c(14.4, 16.8, 11.9, 21.5, 17.2, 18.9, 24.0, 14.8, 12.3, 19.1)
)cities_europe# A tibble: 6 × 2
city population_mio
<chr> <dbl>
1 Berlin 3.9
2 Hamburg 1.9
3 Munich 1.5
4 Copenhagen 0.7
5 Amsterdam 0.9
6 London 9 cities_rent# A tibble: 10 × 2
city rent_sqm
<chr> <dbl>
1 Berlin 18.3
2 Hamburg 17.2
3 Munich 22.6
4 Frankfurt 19.6
5 Cologne 15.2
6 Duesseldorf 16.0
7 Stuttgart 17.3
8 Leipzig 11.4
9 Dresden 7.33
10 Nuremberg 9.65cities_stats# A tibble: 10 × 3
city area_km2 green_space_pct
<chr> <dbl> <dbl>
1 Berlin 892 14.4
2 Hamburg 755 16.8
3 Munich 310 11.9
4 Frankfurt 248 21.5
5 Cologne 405 17.2
6 Duesseldorf 217 18.9
7 Stuttgart 207 24
8 Leipzig 297 14.8
9 Dresden 328 12.3
10 Nuremberg 186 19.1Note that cities_europe contains three German cities (Berlin, Hamburg, Munich) that also appear in the other two tibbles, plus three non-German cities. The tibbles cities_rent and cities_stats have exactly the same ten German cities but different columns.
The Concept: Key Columns
In a join, you specify which column(s) should be used as “keys”. R then searches for matching values in this column and combines the corresponding rows.
In our example data, city is the obvious key column - it appears in all three tibbles and uniquely identifies each row.
Mutating Joins
“Mutating joins” add columns from one table to another - they “mutate” the source table by extending it with new columns. There are four variants that differ in which rows are included in the result.
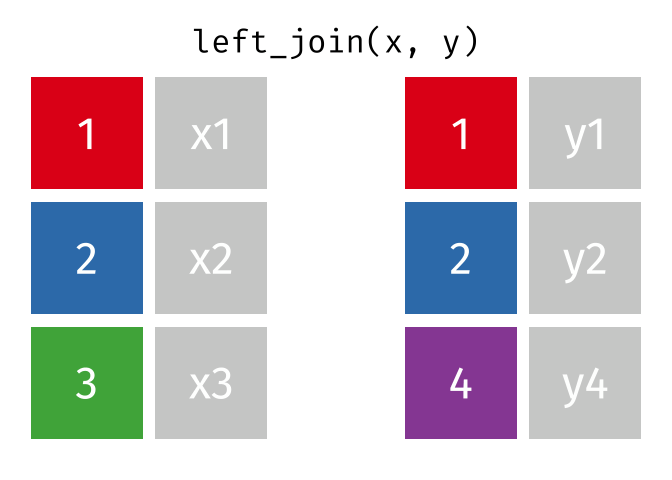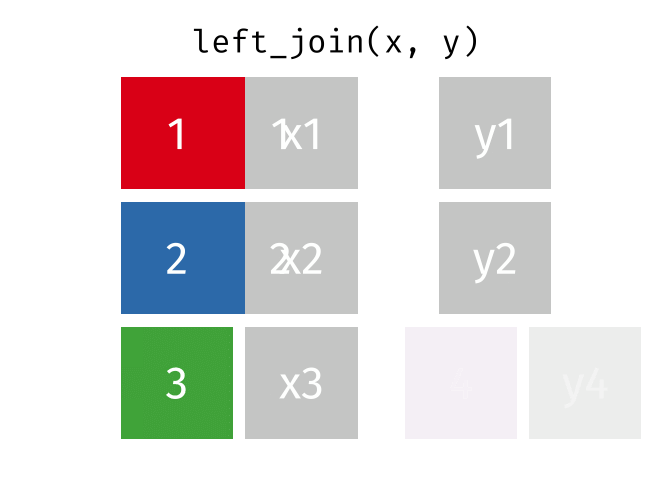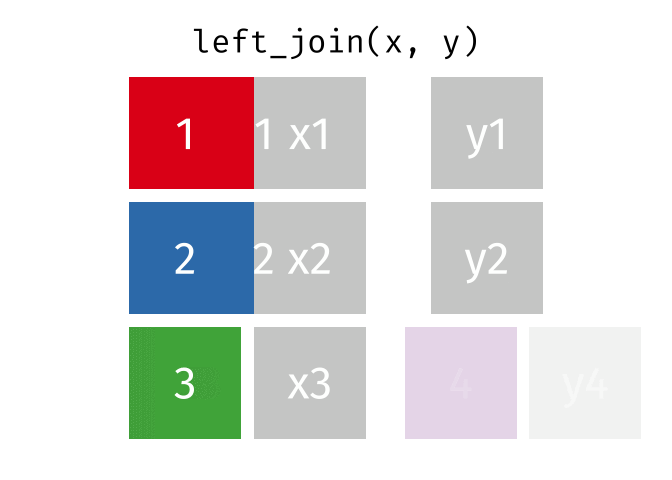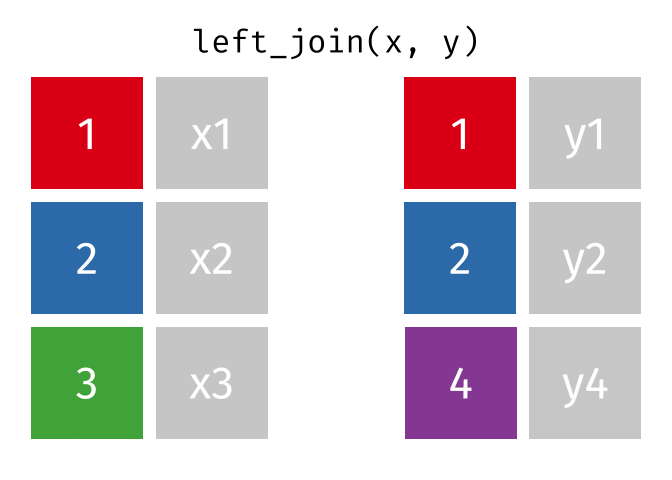
left_join()
The left_join() keeps all rows from the left table and adds matching columns from the right table. If there is no matching partner in the right table, the new columns are filled with NA.
The animated graphics in this chapter come from Garrick Aden-Buie. He has created a fantastic collection of visualizations there that illustrate the different join types and other tidyverse operations. Worth a visit!
# A tibble: 6 × 3
city population_mio rent_sqm
<chr> <dbl> <dbl>
1 Berlin 3.9 18.3
2 Hamburg 1.9 17.2
3 Munich 1.5 22.6
4 Copenhagen 0.7 NA
5 Amsterdam 0.9 NA
6 London 9 NA We can see:
- All 6 cities from
cities_europeare in the result - Berlin, Hamburg, and Munich have received rental prices
- Copenhagen, Amsterdam, and London have
NAforrent_sqmbecause they don’t appear incities_rent
The left_join() is the most commonly used join because you often have a “main table” that you want to extend with additional information without losing rows.
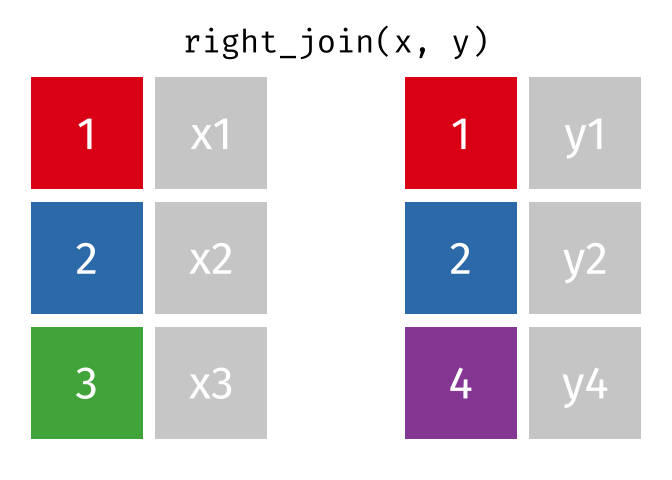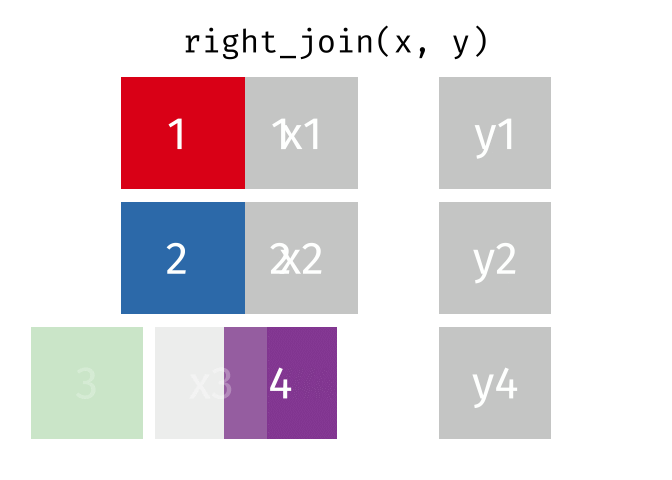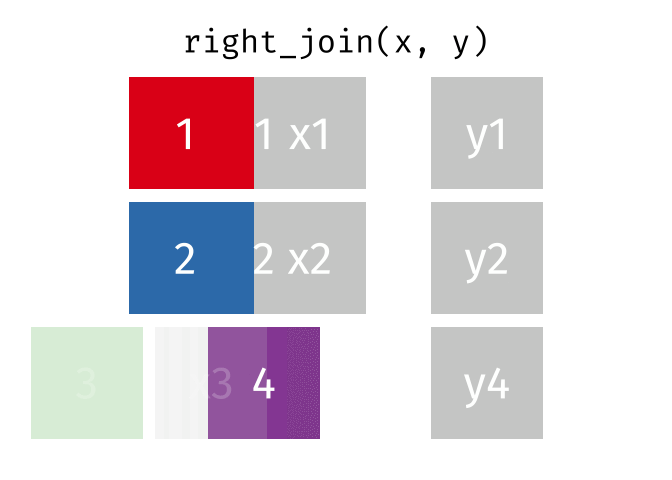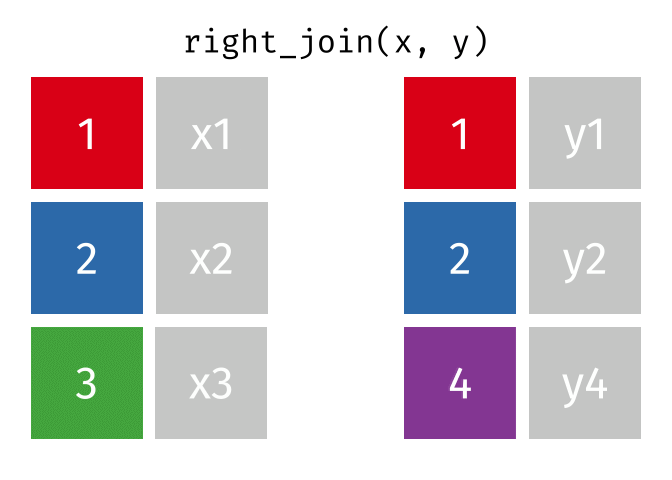
right_join()
The right_join() is the mirror image of left_join(): it keeps all rows from the right table.
cities_europe %>%
right_join(cities_rent, by = "city")# A tibble: 10 × 3
city population_mio rent_sqm
<chr> <dbl> <dbl>
1 Berlin 3.9 18.3
2 Hamburg 1.9 17.2
3 Munich 1.5 22.6
4 Frankfurt NA 19.6
5 Cologne NA 15.2
6 Duesseldorf NA 16.0
7 Stuttgart NA 17.3
8 Leipzig NA 11.4
9 Dresden NA 7.33
10 Nuremberg NA 9.65Now we have:
- All 10 German cities from
cities_rent - Berlin, Hamburg, and Munich have population figures
- The 7 other German cities have
NAforpopulation_mio
In practice, instead of right_join(a, b) you can simply write left_join(b, a) - the result is the same (only the column order differs). Many R users therefore use almost exclusively left_join().
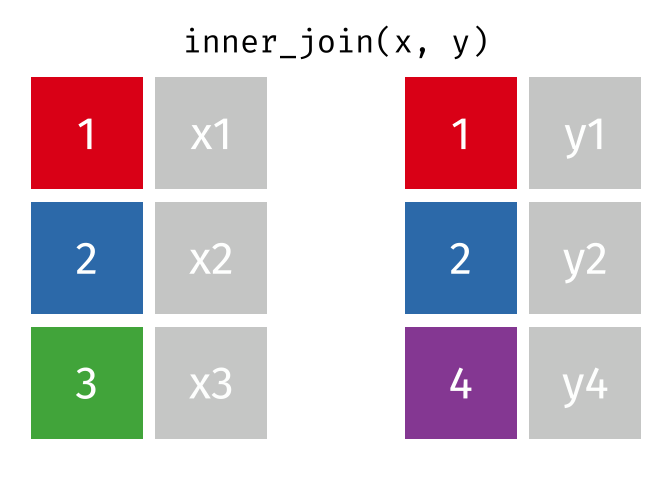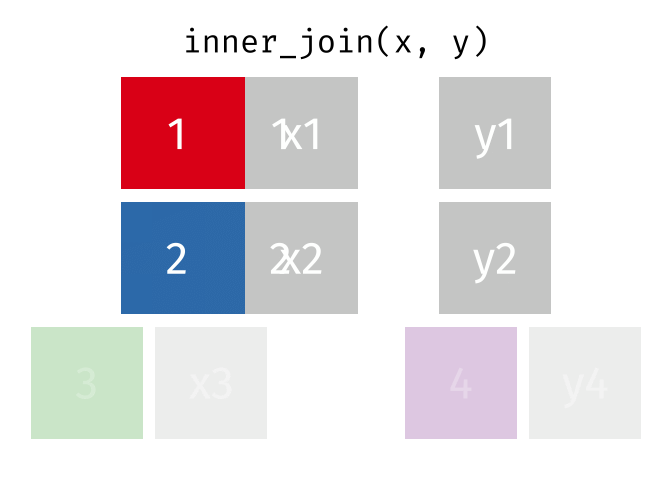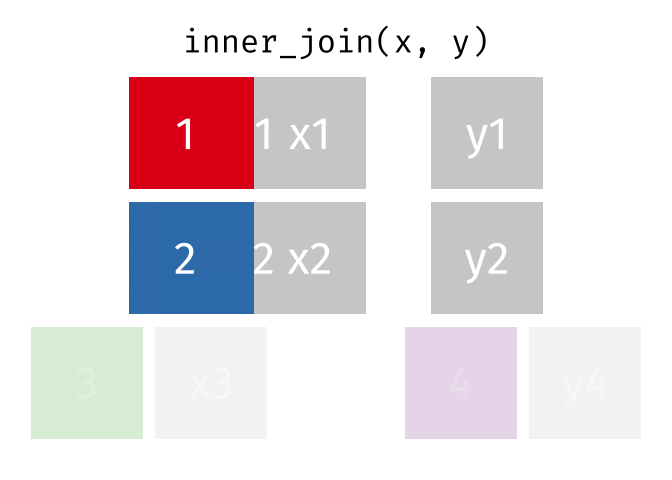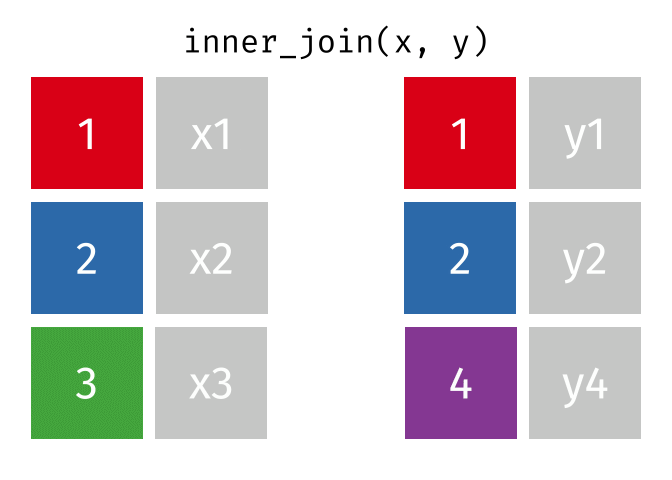
inner_join()
The inner_join() keeps only rows that appear in both tables. Rows without a partner are completely excluded.
cities_europe %>%
inner_join(cities_rent, by = "city")# A tibble: 3 × 3
city population_mio rent_sqm
<chr> <dbl> <dbl>
1 Berlin 3.9 18.3
2 Hamburg 1.9 17.2
3 Munich 1.5 22.6Only Berlin, Hamburg, and Munich remain - the only cities that appear in both tables. There are no NA values in the result.
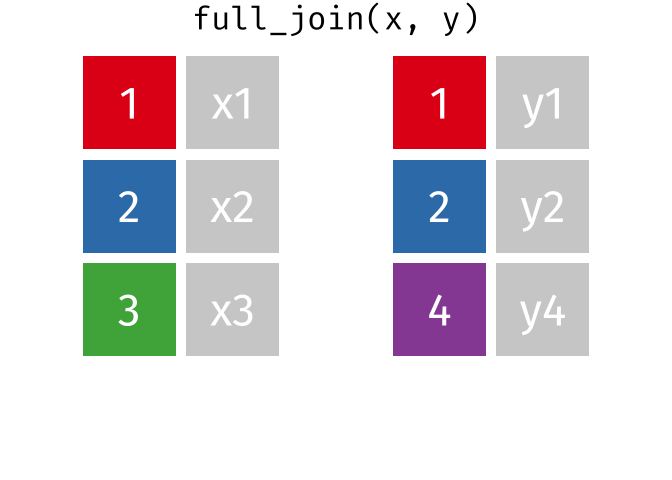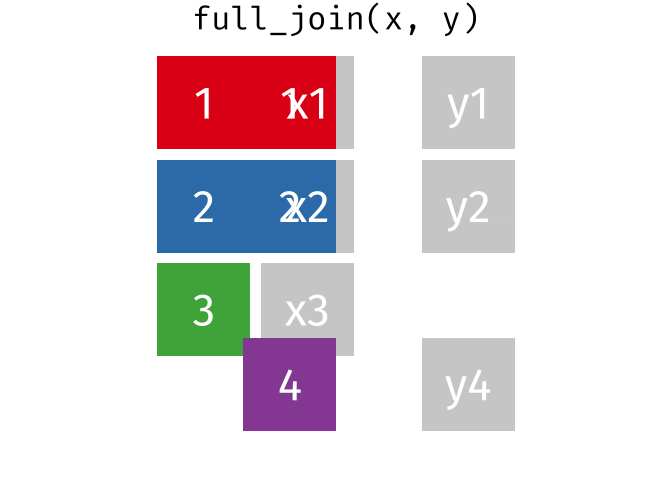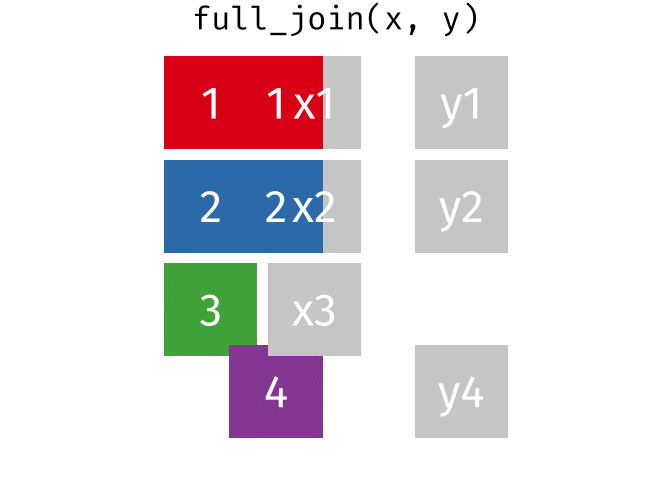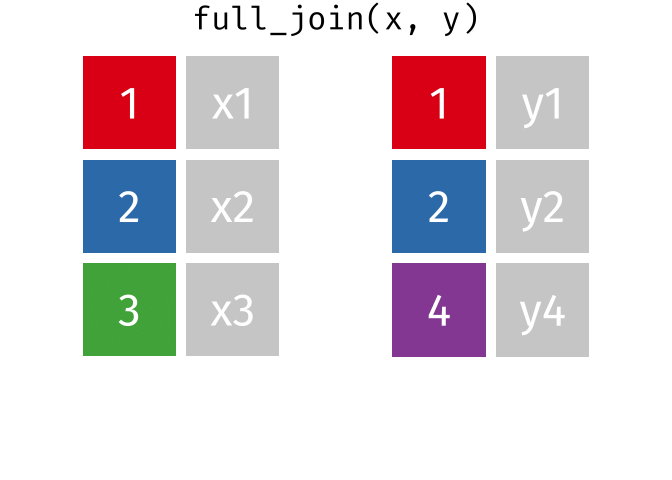
full_join()
The full_join() keeps all rows from both tables. This is the most “generous” variant.
# A tibble: 13 × 3
city population_mio rent_sqm
<chr> <dbl> <dbl>
1 Berlin 3.9 18.3
2 Hamburg 1.9 17.2
3 Munich 1.5 22.6
4 Copenhagen 0.7 NA
5 Amsterdam 0.9 NA
6 London 9 NA
7 Frankfurt NA 19.6
8 Cologne NA 15.2
9 Duesseldorf NA 16.0
10 Stuttgart NA 17.3
11 Leipzig NA 11.4
12 Dresden NA 7.33
13 Nuremberg NA 9.65The result has 13 rows: 3 German cities with complete data, 3 non-German cities (population only), and 7 additional German cities (rent only).
Exercise: Joins with Plant Data
First prepare the data:
# Load and extend PlantGrowth dataset
data(PlantGrowth)
# Dataset 1: Weight measurements with unique ID
plants_weight <- PlantGrowth %>%
mutate(plant_id = 1:n()) %>%
select(plant_id, group, weight)
# Dataset 2: Height measurements (only available for some plants!)
set.seed(123)
plants_height <- tibble(
plant_id = c(1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29),
height_cm = round(rnorm(15, mean = 26, sd = 3), 1)
)
# View the datasets
plants_weight plant_id group weight
1 1 ctrl 4.17
2 2 ctrl 5.58
3 3 ctrl 5.18
4 4 ctrl 6.11
5 5 ctrl 4.50
6 6 ctrl 4.61
7 7 ctrl 5.17
8 8 ctrl 4.53
9 9 ctrl 5.33
10 10 ctrl 5.14
11 11 trt1 4.81
12 12 trt1 4.17
13 13 trt1 4.41
14 14 trt1 3.59
15 15 trt1 5.87
16 16 trt1 3.83
17 17 trt1 6.03
18 18 trt1 4.89
19 19 trt1 4.32
20 20 trt1 4.69
21 21 trt2 6.31
22 22 trt2 5.12
23 23 trt2 5.54
24 24 trt2 5.50
25 25 trt2 5.37
26 26 trt2 5.29
27 27 trt2 4.92
28 28 trt2 6.15
29 29 trt2 5.80
30 30 trt2 5.26plants_height# A tibble: 15 × 2
plant_id height_cm
<dbl> <dbl>
1 1 24.3
2 3 25.3
3 5 30.7
4 7 26.2
5 9 26.4
6 11 31.1
7 13 27.4
8 15 22.2
9 17 23.9
10 19 24.7
11 21 29.7
12 23 27.1
13 25 27.2
14 27 26.3
15 29 24.3Answer the following questions using the appropriate join functions:
Add the height measurements to all plants. Plants without height measurement should get
NA. How many plants have a height measurement?Create a dataset with only the plants for which both weight and height were measured.
Which plants (plant_id) have no height measurement? Use a filtering join.
For the plants with both measurements, calculate the ratio
weight / height_cmand store it in a new columnratio.
plant_id group weight height_cm
1 1 ctrl 4.17 24.3
2 2 ctrl 5.58 NA
3 3 ctrl 5.18 25.3
4 4 ctrl 6.11 NA
5 5 ctrl 4.50 30.7
6 6 ctrl 4.61 NA
7 7 ctrl 5.17 26.2
8 8 ctrl 4.53 NA
9 9 ctrl 5.33 26.4
10 10 ctrl 5.14 NA
11 11 trt1 4.81 31.1
12 12 trt1 4.17 NA
13 13 trt1 4.41 27.4
14 14 trt1 3.59 NA
15 15 trt1 5.87 22.2
16 16 trt1 3.83 NA
17 17 trt1 6.03 23.9
18 18 trt1 4.89 NA
19 19 trt1 4.32 24.7
20 20 trt1 4.69 NA
21 21 trt2 6.31 29.7
22 22 trt2 5.12 NA
23 23 trt2 5.54 27.1
24 24 trt2 5.50 NA
25 25 trt2 5.37 27.2
26 26 trt2 5.29 NA
27 27 trt2 4.92 26.3
28 28 trt2 6.15 NA
29 29 trt2 5.80 24.3
30 30 trt2 5.26 NA[1] 15# b) inner_join: Only plants with both measurements
plants_both <- plants_weight %>%
inner_join(plants_height, by = "plant_id")
plants_both plant_id group weight height_cm
1 1 ctrl 4.17 24.3
2 3 ctrl 5.18 25.3
3 5 ctrl 4.50 30.7
4 7 ctrl 5.17 26.2
5 9 ctrl 5.33 26.4
6 11 trt1 4.81 31.1
7 13 trt1 4.41 27.4
8 15 trt1 5.87 22.2
9 17 trt1 6.03 23.9
10 19 trt1 4.32 24.7
11 21 trt2 6.31 29.7
12 23 trt2 5.54 27.1
13 25 trt2 5.37 27.2
14 27 trt2 4.92 26.3
15 29 trt2 5.80 24.3 plant_id group weight
1 2 ctrl 5.58
2 4 ctrl 6.11
3 6 ctrl 4.61
4 8 ctrl 4.53
5 10 ctrl 5.14
6 12 trt1 4.17
7 14 trt1 3.59
8 16 trt1 3.83
9 18 trt1 4.89
10 20 trt1 4.69
11 22 trt2 5.12
12 24 trt2 5.50
13 26 trt2 5.29
14 28 trt2 6.15
15 30 trt2 5.26 plant_id group weight height_cm ratio
1 1 ctrl 4.17 24.3 0.1716049
2 3 ctrl 5.18 25.3 0.2047431
3 5 ctrl 4.50 30.7 0.1465798
4 7 ctrl 5.17 26.2 0.1973282
5 9 ctrl 5.33 26.4 0.2018939
6 11 trt1 4.81 31.1 0.1546624
7 13 trt1 4.41 27.4 0.1609489
8 15 trt1 5.87 22.2 0.2644144
9 17 trt1 6.03 23.9 0.2523013
10 19 trt1 4.32 24.7 0.1748988
11 21 trt2 6.31 29.7 0.2124579
12 23 trt2 5.54 27.1 0.2044280
13 25 trt2 5.37 27.2 0.1974265
14 27 trt2 4.92 26.3 0.1870722
15 29 trt2 5.80 24.3 0.2386831Different Column Names
Sometimes the key column has different names in the two tables. You can specify this in the by argument:
# A tibble: 10 × 3
city rent_sqm population
<chr> <dbl> <dbl>
1 Berlin 18.3 3.8
2 Hamburg 17.2 1.9
3 Munich 22.6 1.5
4 Frankfurt 19.6 NA
5 Cologne 15.2 NA
6 Duesseldorf 16.0 NA
7 Stuttgart 17.3 NA
8 Leipzig 11.4 NA
9 Dresden 7.33 NA
10 Nuremberg 9.65 NA The syntax by = c("city" = "stadt") means: “Link the city column from the left table with the stadt column from the right table.”
Filtering Joins
Unlike mutating joins, filtering joins do not add new columns. They only filter the rows of the left table based on whether there is a partner in the right table.
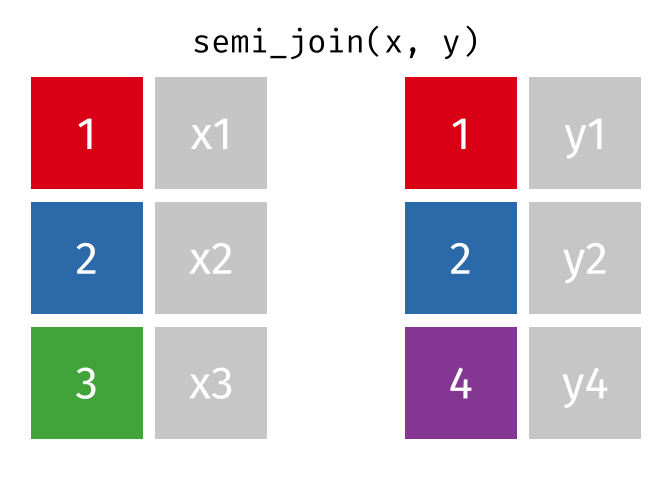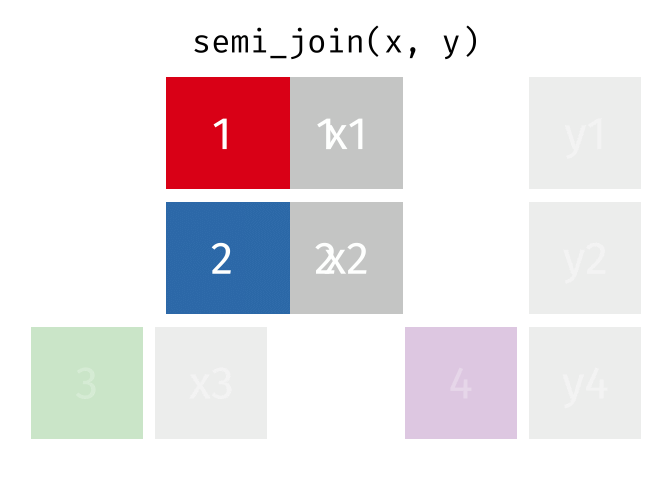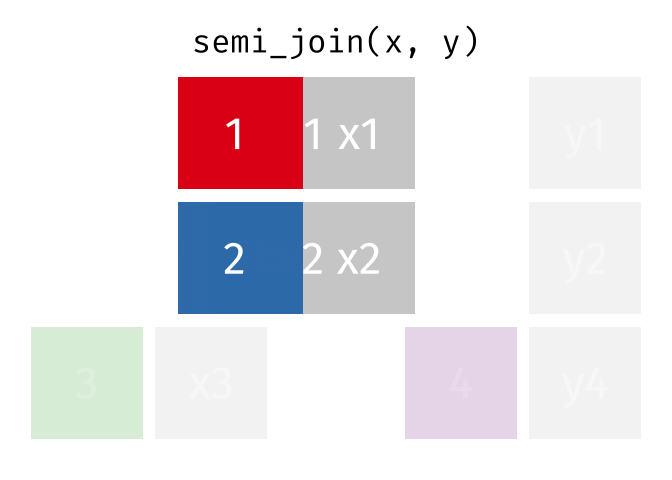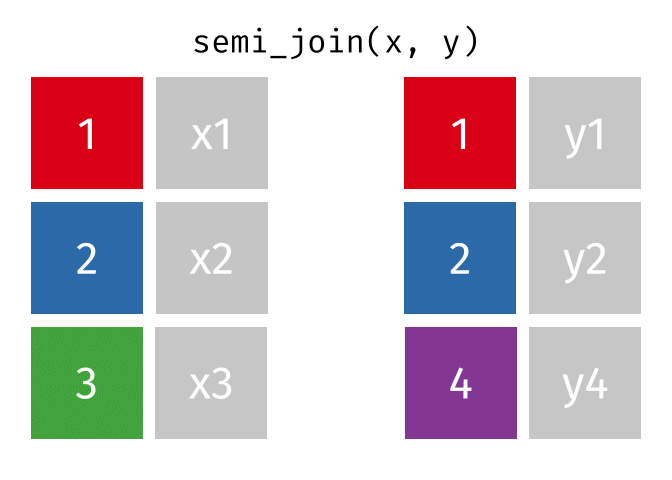
semi_join()
The semi_join() keeps all rows from the left table that have a partner in the right table.
# A tibble: 3 × 2
city population_mio
<chr> <dbl>
1 Berlin 3.9
2 Hamburg 1.9
3 Munich 1.5The result contains only Berlin, Hamburg, and Munich - the European cities for which we have rental data. But: no columns from cities_rent were added! The result only has the columns from cities_europe.
The semi_join() answers the question: “Which rows from table A have a partner in table B?”
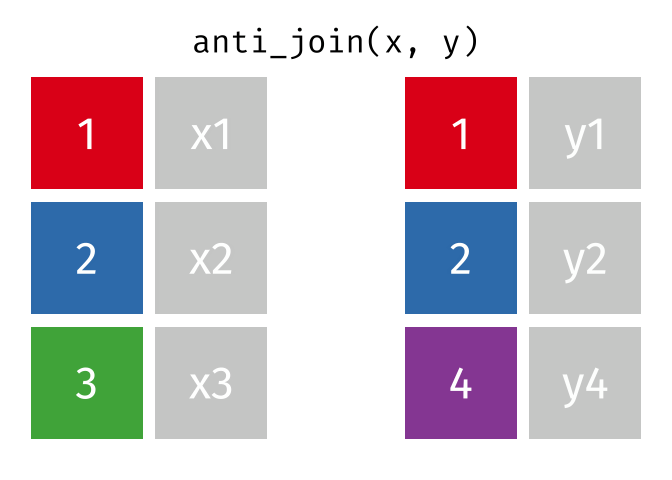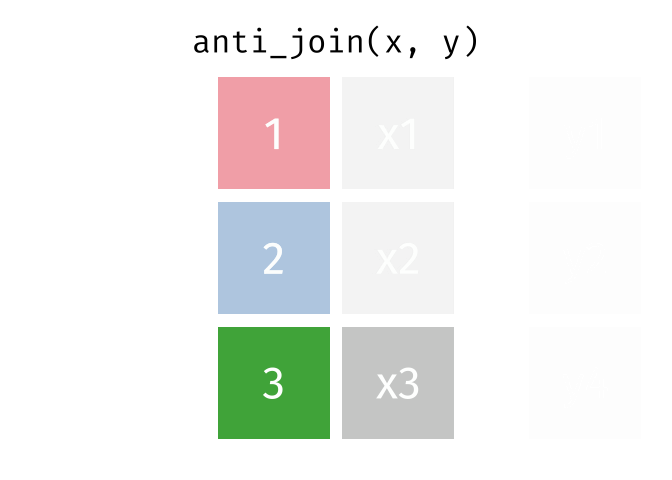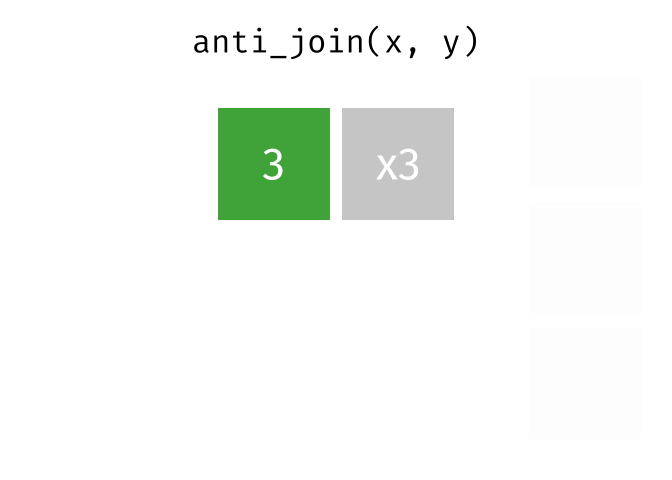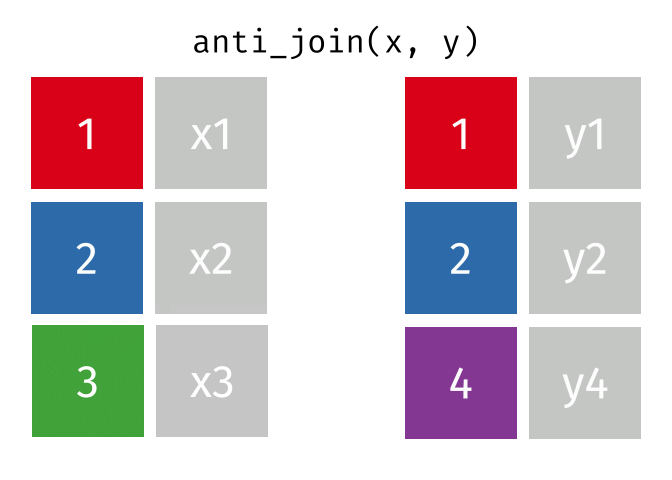
anti_join()
The anti_join() is the opposite: it keeps all rows from the left table that have no partner in the right table.
# A tibble: 3 × 2
city population_mio
<chr> <dbl>
1 Copenhagen 0.7
2 Amsterdam 0.9
3 London 9 Copenhagen, Amsterdam, and London - the European cities for which we have no rental data.
The anti_join() is very useful for data quality checks: “Which records are missing?” or “Which IDs from system A don’t exist in system B?”
Set Operations
Set operations treat tables as mathematical sets. They only work when both tables have exactly the same columns. They then compare entire rows (not individual key columns).
For the examples, we create two small tables with identical columns:
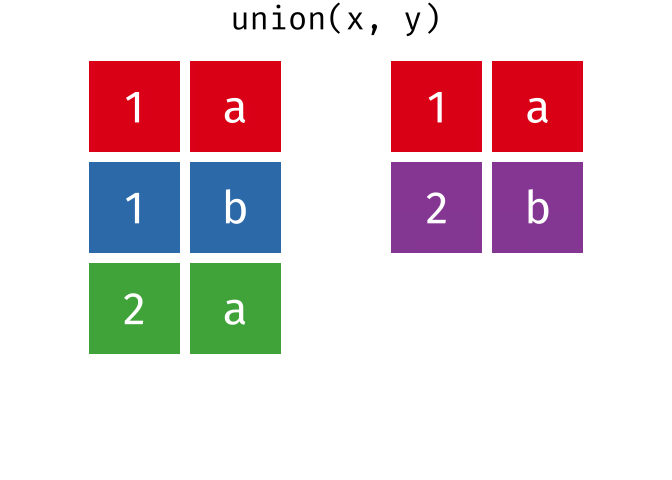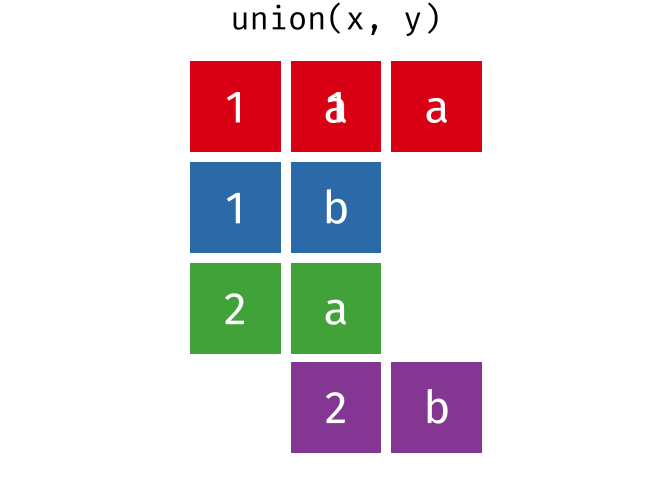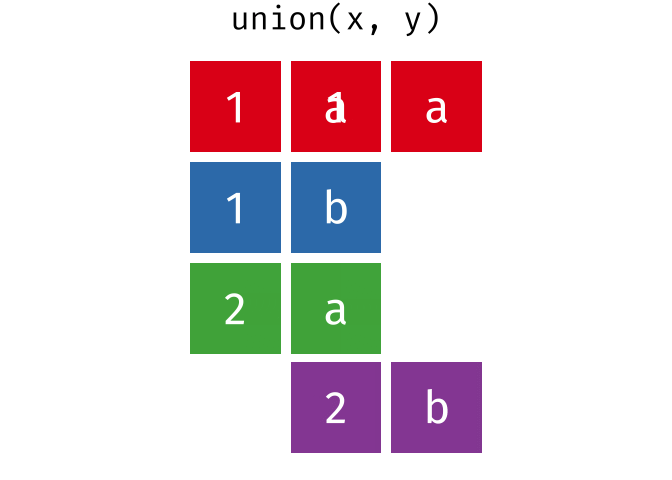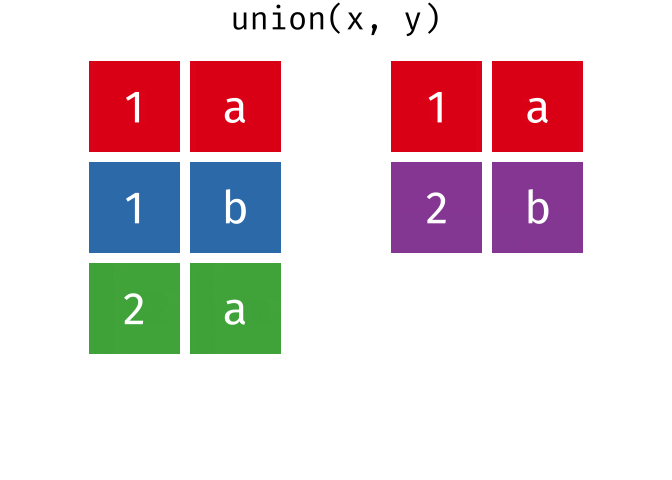
union()
union() returns all unique rows from both tables - the union set.
union(set_a, set_b)# A tibble: 4 × 2
city country
<chr> <chr>
1 Berlin Germany
2 Hamburg Germany
3 Munich Germany
4 Frankfurt GermanyHamburg and Munich appear in both tables but appear only once in the result.
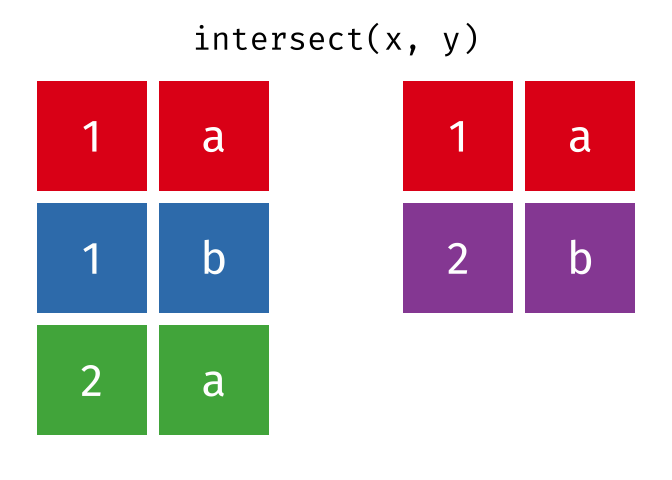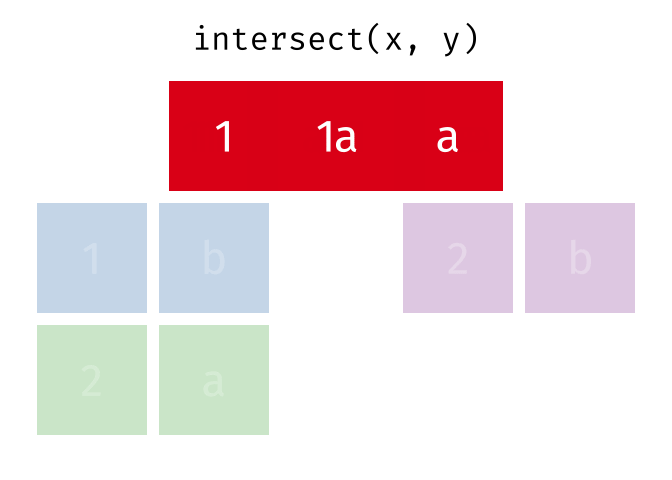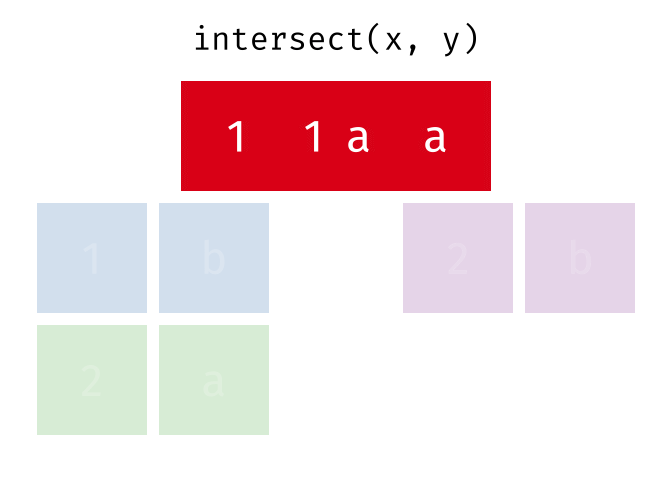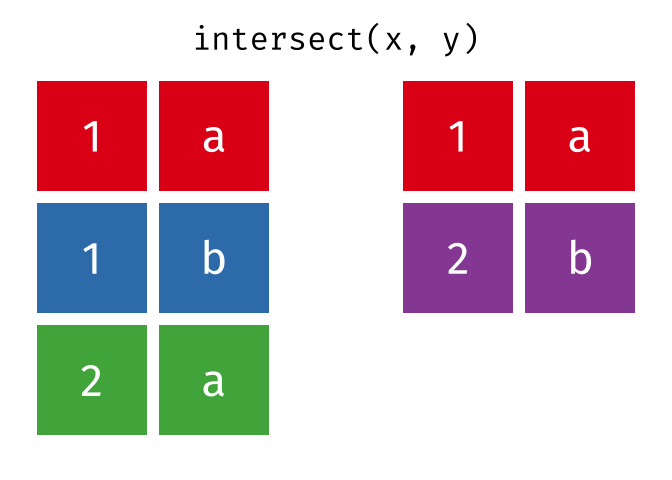
intersect()
intersect() returns only the rows that appear in both tables - the intersection.
intersect(set_a, set_b)# A tibble: 2 × 2
city country
<chr> <chr>
1 Hamburg Germany
2 Munich GermanyOnly Hamburg and Munich are in both tables.
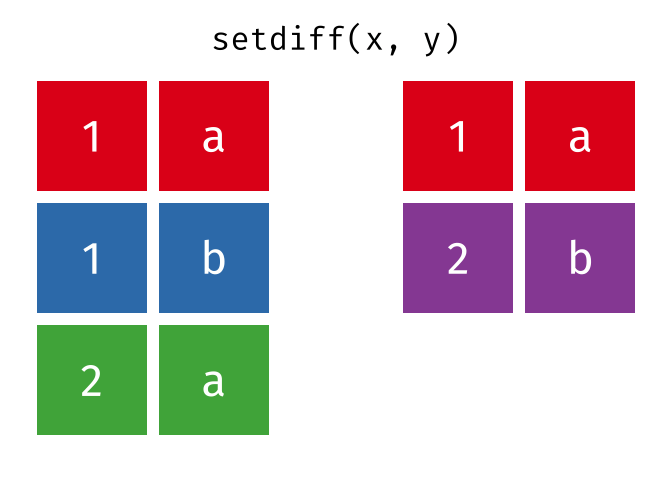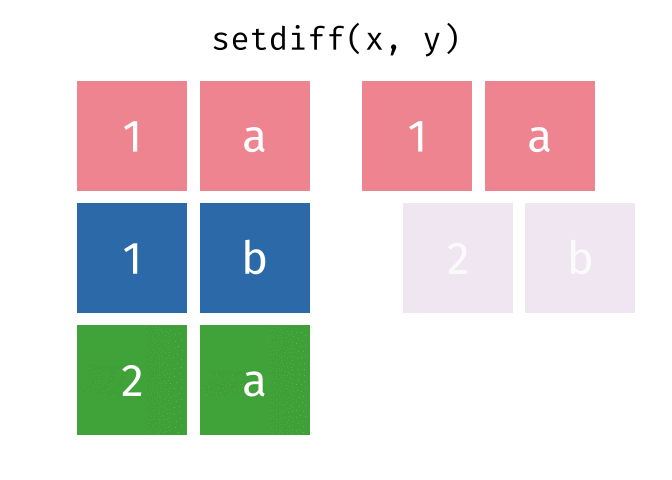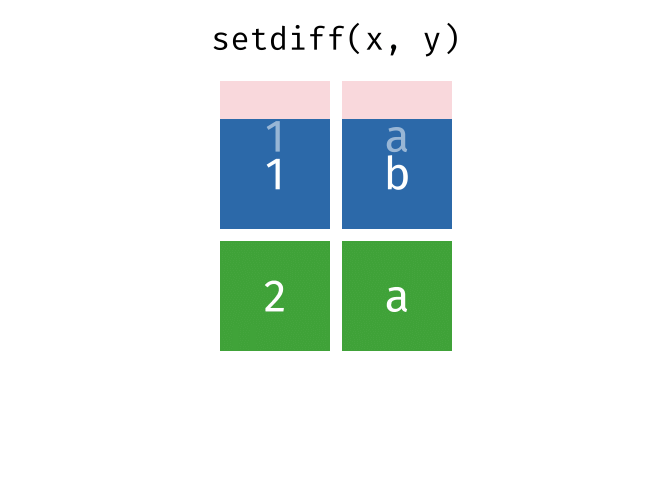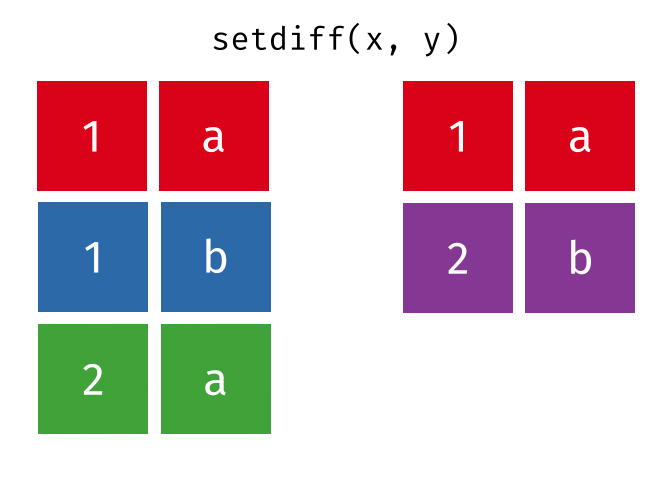
setdiff()
setdiff() returns the rows that are in the first but not in the second table - the difference set.
setdiff(set_a, set_b)# A tibble: 1 × 2
city country
<chr> <chr>
1 Berlin GermanyBerlin is only in set_a.
Reshaping Data (Wide ↔︎ Long)
Often we need to transform data between two formats:
- Wide format: Each variable has its own column
- Long format: Variable names become values in a column
Which format is “correct” depends on the use case. For many tidyverse functions and ggplot2, the long format is better suited, while the wide format is often more readable for humans.
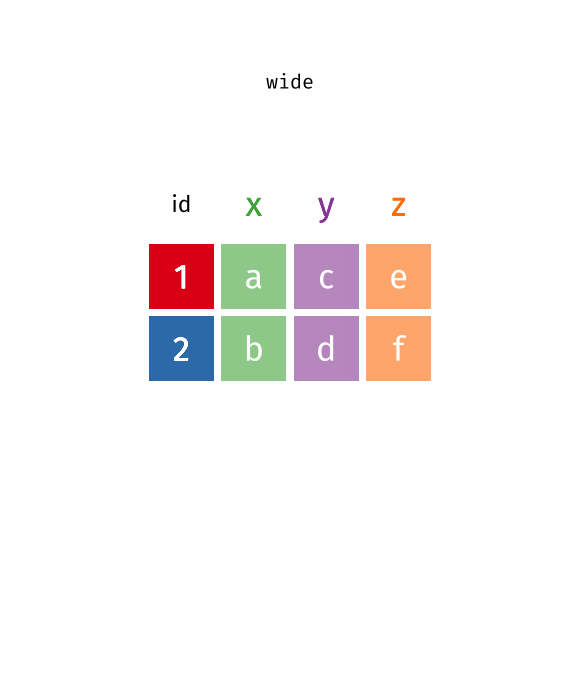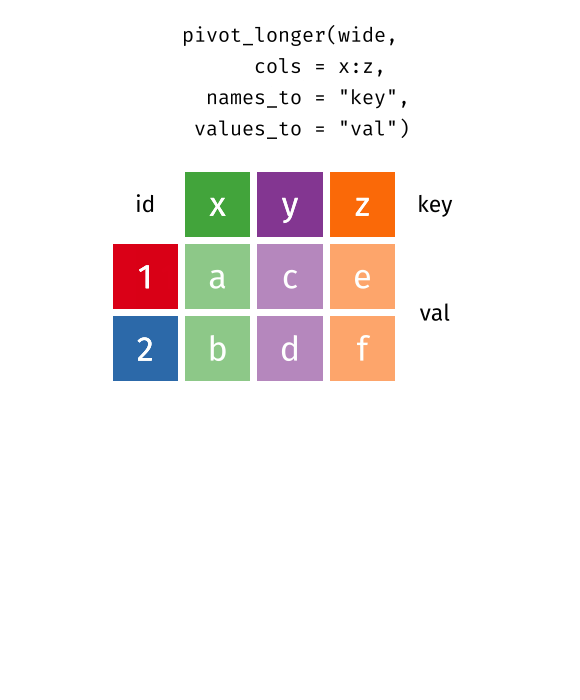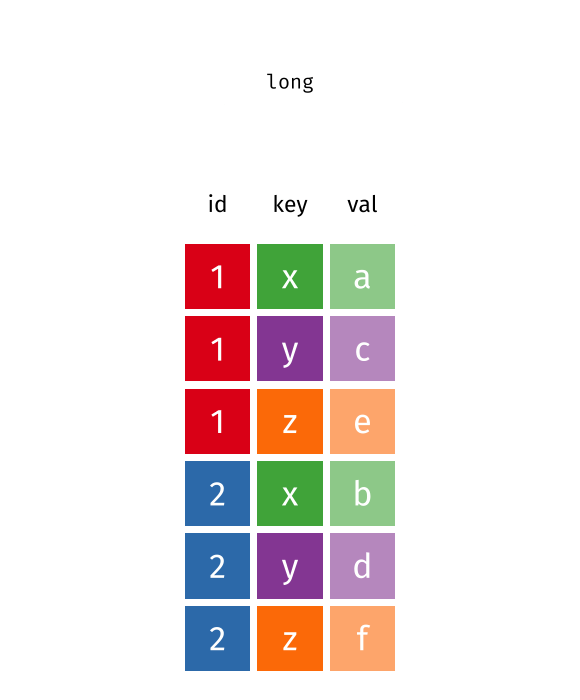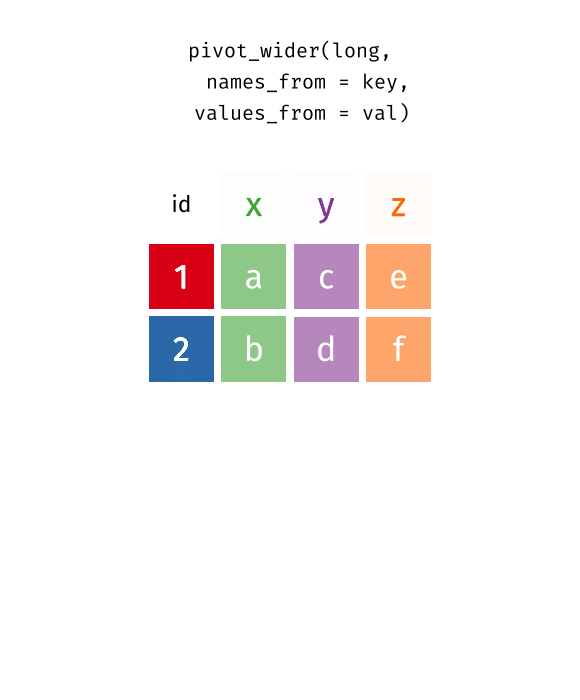
pivot_longer()
pivot_longer() transforms data from wide to long format - it makes the table “longer” (more rows, fewer columns).
Let’s look at cities_stats:
cities_stats# A tibble: 10 × 3
city area_km2 green_space_pct
<chr> <dbl> <dbl>
1 Berlin 892 14.4
2 Hamburg 755 16.8
3 Munich 310 11.9
4 Frankfurt 248 21.5
5 Cologne 405 17.2
6 Duesseldorf 217 18.9
7 Stuttgart 207 24
8 Leipzig 297 14.8
9 Dresden 328 12.3
10 Nuremberg 186 19.1This is a typical wide format: each metric (area, green space) has its own column. For some analyses or visualizations, we want to convert this to long format:
cities_stats %>%
pivot_longer(
cols = c(area_km2, green_space_pct),
names_to = "metric",
values_to = "value"
)# A tibble: 20 × 3
city metric value
<chr> <chr> <dbl>
1 Berlin area_km2 892
2 Berlin green_space_pct 14.4
3 Hamburg area_km2 755
4 Hamburg green_space_pct 16.8
5 Munich area_km2 310
6 Munich green_space_pct 11.9
7 Frankfurt area_km2 248
8 Frankfurt green_space_pct 21.5
9 Cologne area_km2 405
10 Cologne green_space_pct 17.2
11 Duesseldorf area_km2 217
12 Duesseldorf green_space_pct 18.9
13 Stuttgart area_km2 207
14 Stuttgart green_space_pct 24
15 Leipzig area_km2 297
16 Leipzig green_space_pct 14.8
17 Dresden area_km2 328
18 Dresden green_space_pct 12.3
19 Nuremberg area_km2 186
20 Nuremberg green_space_pct 19.1The key arguments:
-
cols: Which columns should be “collapsed”? -
names_to: What should the new column be called that contains the old column names? -
values_to: What should the new column be called that contains the values?
Now each city has two rows - one per metric. This is ideal for ggplot2 when you want to display both metrics in a faceted plot, for example.
Column Selection with Helper Functions
Instead of listing columns individually, you can use helper functions:
# All columns except "city"
cities_stats %>%
pivot_longer(
cols = -city,
names_to = "metric",
values_to = "value"
)# A tibble: 20 × 3
city metric value
<chr> <chr> <dbl>
1 Berlin area_km2 892
2 Berlin green_space_pct 14.4
3 Hamburg area_km2 755
4 Hamburg green_space_pct 16.8
5 Munich area_km2 310
6 Munich green_space_pct 11.9
7 Frankfurt area_km2 248
8 Frankfurt green_space_pct 21.5
9 Cologne area_km2 405
10 Cologne green_space_pct 17.2
11 Duesseldorf area_km2 217
12 Duesseldorf green_space_pct 18.9
13 Stuttgart area_km2 207
14 Stuttgart green_space_pct 24
15 Leipzig area_km2 297
16 Leipzig green_space_pct 14.8
17 Dresden area_km2 328
18 Dresden green_space_pct 12.3
19 Nuremberg area_km2 186
20 Nuremberg green_space_pct 19.1# All numeric columns
cities_stats %>%
pivot_longer(
cols = where(is.numeric),
names_to = "metric",
values_to = "value"
)# A tibble: 20 × 3
city metric value
<chr> <chr> <dbl>
1 Berlin area_km2 892
2 Berlin green_space_pct 14.4
3 Hamburg area_km2 755
4 Hamburg green_space_pct 16.8
5 Munich area_km2 310
6 Munich green_space_pct 11.9
7 Frankfurt area_km2 248
8 Frankfurt green_space_pct 21.5
9 Cologne area_km2 405
10 Cologne green_space_pct 17.2
11 Duesseldorf area_km2 217
12 Duesseldorf green_space_pct 18.9
13 Stuttgart area_km2 207
14 Stuttgart green_space_pct 24
15 Leipzig area_km2 297
16 Leipzig green_space_pct 14.8
17 Dresden area_km2 328
18 Dresden green_space_pct 12.3
19 Nuremberg area_km2 186
20 Nuremberg green_space_pct 19.1pivot_wider()
pivot_wider() is the inverse function: it transforms from long to wide format - the table becomes “wider” (fewer rows, more columns).
First, let’s create a long-format table:
cities_long <- cities_stats %>%
pivot_longer(
cols = -city,
names_to = "metric",
values_to = "value"
)
cities_long# A tibble: 20 × 3
city metric value
<chr> <chr> <dbl>
1 Berlin area_km2 892
2 Berlin green_space_pct 14.4
3 Hamburg area_km2 755
4 Hamburg green_space_pct 16.8
5 Munich area_km2 310
6 Munich green_space_pct 11.9
7 Frankfurt area_km2 248
8 Frankfurt green_space_pct 21.5
9 Cologne area_km2 405
10 Cologne green_space_pct 17.2
11 Duesseldorf area_km2 217
12 Duesseldorf green_space_pct 18.9
13 Stuttgart area_km2 207
14 Stuttgart green_space_pct 24
15 Leipzig area_km2 297
16 Leipzig green_space_pct 14.8
17 Dresden area_km2 328
18 Dresden green_space_pct 12.3
19 Nuremberg area_km2 186
20 Nuremberg green_space_pct 19.1Now we transform back to wide format:
cities_long %>%
pivot_wider(
names_from = metric,
values_from = value
)# A tibble: 10 × 3
city area_km2 green_space_pct
<chr> <dbl> <dbl>
1 Berlin 892 14.4
2 Hamburg 755 16.8
3 Munich 310 11.9
4 Frankfurt 248 21.5
5 Cologne 405 17.2
6 Duesseldorf 217 18.9
7 Stuttgart 207 24
8 Leipzig 297 14.8
9 Dresden 328 12.3
10 Nuremberg 186 19.1The key arguments:
-
names_from: Which column contains the future column names? -
values_from: Which column contains the values?
You may have already used other functions in this context. Here are some alternatives, some of which are now deprecated:
Typical Use Case: Cross Tables
pivot_wider() is also useful for creating cross tables. Suppose we have sales data:
# A tibble: 4 × 3
product quarter revenue
<chr> <chr> <dbl>
1 Apple Q1 100
2 Apple Q2 120
3 Pear Q1 80
4 Pear Q2 90sales %>%
pivot_wider(
names_from = quarter,
values_from = revenue
)# A tibble: 2 × 3
product Q1 Q2
<chr> <dbl> <dbl>
1 Apple 100 120
2 Pear 80 90Now we have a clear cross table with products in rows and quarters in columns.
Exercise: Pivoting Workflow
First prepare a dataset in long format:
# Simulate PlantGrowth with multiple measurements
set.seed(42)
plants_long <- PlantGrowth %>%
mutate(
plant_id = 1:n(),
height_cm = weight * 5 + rnorm(n(), mean = 0, sd = 2)
) %>%
pivot_longer(
cols = c(weight, height_cm),
names_to = "measurement",
values_to = "value"
) %>%
select(plant_id, group, measurement, value)
plants_long# A tibble: 60 × 4
plant_id group measurement value
<int> <fct> <chr> <dbl>
1 1 ctrl weight 4.17
2 1 ctrl height_cm 23.6
3 2 ctrl weight 5.58
4 2 ctrl height_cm 26.8
5 3 ctrl weight 5.18
6 3 ctrl height_cm 26.6
7 4 ctrl weight 6.11
8 4 ctrl height_cm 31.8
9 5 ctrl weight 4.5
10 5 ctrl height_cm 23.3
# ℹ 50 more rowsPerform the following transformations:
Transform
plants_longto wide format so thatweightandheight_cmeach have their own columns.Add a new column
bmi(Body Mass Index for plants) that calculates the ratioweight / height_cm.Transform the dataset back to long format so that all three variables (
weight,height_cm, andbmi) appear in themeasurementcolumn.
# a) Create wide format
plants_wide <- plants_long %>%
pivot_wider(
names_from = measurement,
values_from = value
)
plants_wide# A tibble: 30 × 4
plant_id group weight height_cm
<int> <fct> <dbl> <dbl>
1 1 ctrl 4.17 23.6
2 2 ctrl 5.58 26.8
3 3 ctrl 5.18 26.6
4 4 ctrl 6.11 31.8
5 5 ctrl 4.5 23.3
6 6 ctrl 4.61 22.8
7 7 ctrl 5.17 28.9
8 8 ctrl 4.53 22.5
9 9 ctrl 5.33 30.7
10 10 ctrl 5.14 25.6
# ℹ 20 more rows# A tibble: 30 × 5
plant_id group weight height_cm bmi
<int> <fct> <dbl> <dbl> <dbl>
1 1 ctrl 4.17 23.6 0.177
2 2 ctrl 5.58 26.8 0.208
3 3 ctrl 5.18 26.6 0.195
4 4 ctrl 6.11 31.8 0.192
5 5 ctrl 4.5 23.3 0.193
6 6 ctrl 4.61 22.8 0.202
7 7 ctrl 5.17 28.9 0.179
8 8 ctrl 4.53 22.5 0.202
9 9 ctrl 5.33 30.7 0.174
10 10 ctrl 5.14 25.6 0.201
# ℹ 20 more rows# c) Back to long format (all three variables)
plants_final_long <- plants_wide %>%
pivot_longer(
cols = c(weight, height_cm, bmi),
names_to = "measurement",
values_to = "value"
)
plants_final_long# A tibble: 90 × 4
plant_id group measurement value
<int> <fct> <chr> <dbl>
1 1 ctrl weight 4.17
2 1 ctrl height_cm 23.6
3 1 ctrl bmi 0.177
4 2 ctrl weight 5.58
5 2 ctrl height_cm 26.8
6 2 ctrl bmi 0.208
7 3 ctrl weight 5.18
8 3 ctrl height_cm 26.6
9 3 ctrl bmi 0.195
10 4 ctrl weight 6.11
# ℹ 80 more rowsSummary
Well done! You now master the most important techniques for combining and reshaping tables in R.
-
Stacking Tables:
-
bind_rows(): Stack rows vertically - works even with different columns (missing ones are filled with NA) -
bind_cols(): Glue columns horizontally - Caution: no intelligent linking, order must match!
-
-
Mutating Joins (add columns):
-
left_join(): Keep all rows from the left table - the default case -
right_join(): Keep all rows from the right table -
inner_join(): Only rows with a partner in both tables -
full_join(): All rows from both tables
-
-
Filtering Joins (only filter, no new columns):
-
semi_join(): Rows from x that have a partner in y -
anti_join(): Rows from x that have no partner in y - ideal for “What’s missing?” questions
-
-
Set Operations (tables as sets, require identical columns):
-
union(): All unique rows from both -
intersect(): Only rows that appear in both -
setdiff(): Rows from x that are not in y
-
-
Pivoting (change data format):
-
pivot_longer(): Wide → Long (more rows, fewer columns) -
pivot_wider(): Long → Wide (fewer rows, more columns)
-
-
Best Practices:
- For different column names:
by = c("name_left" = "name_right") - When in doubt, use
left_join()instead ofbind_cols() - Use
anti_join()for data quality checks
- For different column names:
Citation
@online{schmidt2026,
author = {{Dr. Paul Schmidt}},
publisher = {BioMath GmbH},
title = {1. {Combining} {Tables}},
date = {2026-02-07},
url = {https://biomathcontent.netlify.app/content/r_more/01_joins.html},
langid = {en}
}